

With the growing power of large language models (LLMs), structured healthcare data like FHIR (Fast Healthcare Interoperability Resources) is becoming increasingly vital. FHIR is the standard for health data exchange in the U.S. and is widely accepted in clinical systems due to its interoperability and detailed data structure. In the context of AI, especially in Retrieval-Augmented Generation (RAG) systems, FHIR data allows healthcare solutions to provide personalized insights about patient lab results, diagnoses, and medical history.
At the ML team, we explored the huge potential of merging FHIR into RAG systems through different strategies we'll share in this blog. We’ll also cover:
- Key challenges in integrating FHIR data
- Strategies for optimizing FHIR data for RAG systems
- Experimental results from different approaches
- Future optimizations to further enhance these systems
The Potential of Combining RAG with FHIR Data
RAG systems can leverage FHIR data to address patient-specific queries effectively. By processing structured data, they generate precise answers that complement the insights provided by specialists, reducing the ambiguity often present in traditional interpretations. For instance, patients can ask, "What do my glucose levels mean?" and receive a tailored response based on their health records, providing clarity on diagnostics and treatment plans.

By combining RAGs and FHIR, users get explanations for complex terms, compare previous results with current ones, and better understand their diagnoses and treatments. It acts as a complement that facilitates data comprehension and enhances communication between patients and healthcare providers.
Key Challenges of Integrating FHIR Data
Despite the potential of combining FHIR with RAG models, there are specific technical challenges:
- Data Complexity: FHIR data, typically stored in JSON format, is hierarchical and granular. Large Language Models (LLMs) work best with plain text, making the conversion from structured JSON to a flat, readable format essential but not straightforward. This complexity presents a challenge when storing and processing FHIR data in vector databases, which are critical for effective performance in RAG systems.
- Relevance: Retrieving the most relevant data points from a complex resource like FHIR can be challenging, especially when the data is fragmented across multiple fields.
Optimizing FHIR Data for RAG Systems
To tackle these challenges, several strategies have been tested to improve data retrieval and make FHIR data more accessible for AI systems:
-
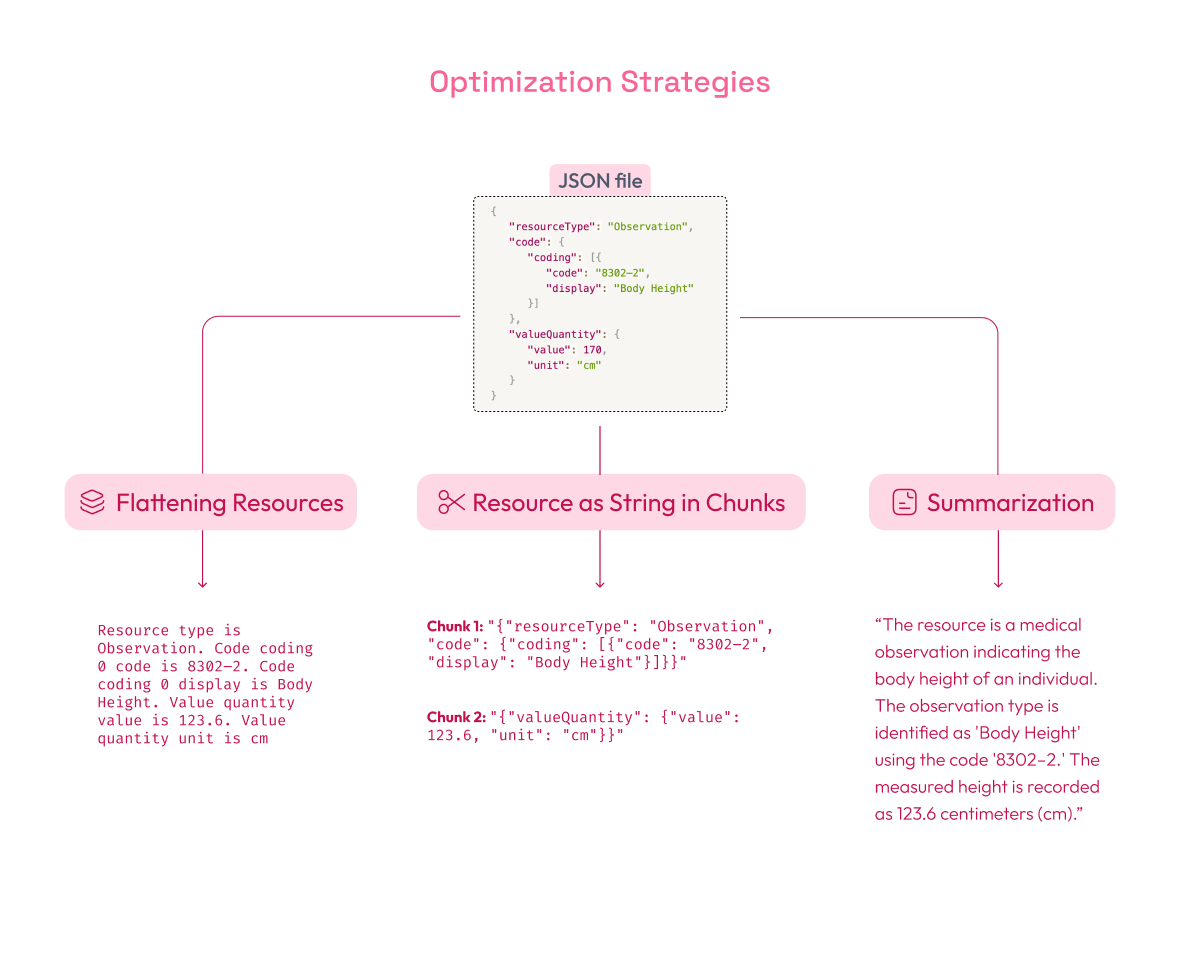
Flattening Resources: This strategy involves converting complex FHIR JSON data into simpler, plain text phrases. The goal of flattening is to remove unnecessary layers of structure (like nested fields) and make the information easier to search.
Example: Suppose you have an observation resource in JSON format:
{
"resourceType": "Observation",
"code": {
"coding": [{
"code": "8302–2",
"display": "Body Height"
}]
},
"valueQuantity": {
"value": 170,
"unit": "cm"
}
}
After flattening, it would be simplified to something like:
"Resource type is Observation. Code coding 0 code is 8302–2. Code coding 0 display is Body Height. Value quantity value is 123.6. Value quantity unit is cm.".
This makes it much easier for a search engine to locate relevant concepts without navigating through nested fields.
-
Resource as String in Chunks: In this method, FHIR resources are broken into smaller, text-based chunks. A chunk is a section of data that can be processed individually, usually a fragment of the original resource. By segmenting the resource, the RAG model can focus on relevant data sections rather than processing the entire resource at once.
Example: If you break the JSON into chunks, you might have:
- Chunk 1:
"{"resourceType": "Observation", "code": {"coding": [{"code": "8302–2", "display": "Body Height"}]}}" - Chunk 2:
"{"valueQuantity": {"value": 123.6, "unit": "cm"}}"
Each chunk is then treated as an independent unit, making retrieving precise information easier for the model.
- Chunk 1:
-
Summarization: Using LLMs, each FHIR resource can be summarized into concise blocks of text (e.g., 800 characters) that capture its key elements. Summaries help improve retrieval precision by focusing on the most important information.
For example, for the FHIR resource we are using, the summary could be:
“The resource is a medical observation indicating the body height of an individual. The observation type is identified as 'Body Height' using the code '8302–2.' The measured height is recorded as 123.6 centimeters (cm).”
Experimental Results Using Elasticsearch
We evaluated these strategies using Elasticsearch with boosting for text and cosine similarity to retrieve relevant FHIR data. The key metrics used were:
- Retrieval Accuracy: The percentage of queries where at least one relevant chunk was retrieved, showing how often the system found correct information.
- MRR (Mean Reciprocal Rank): Measures the ranking position of the first relevant chunk in the results. Higher values indicate that relevant information appears earlier (higher in the list), which improves the system’s effectiveness in presenting important data at the top.
In some experiments, ID and Date were included in the queries. These represent unique identifiers such as resource IDs or dates of observations, which are key fields in FHIR resources. Including them helps reduce search ambiguity, as they provide precise markers that help the system target the correct resource more effectively.
Here are the findings:
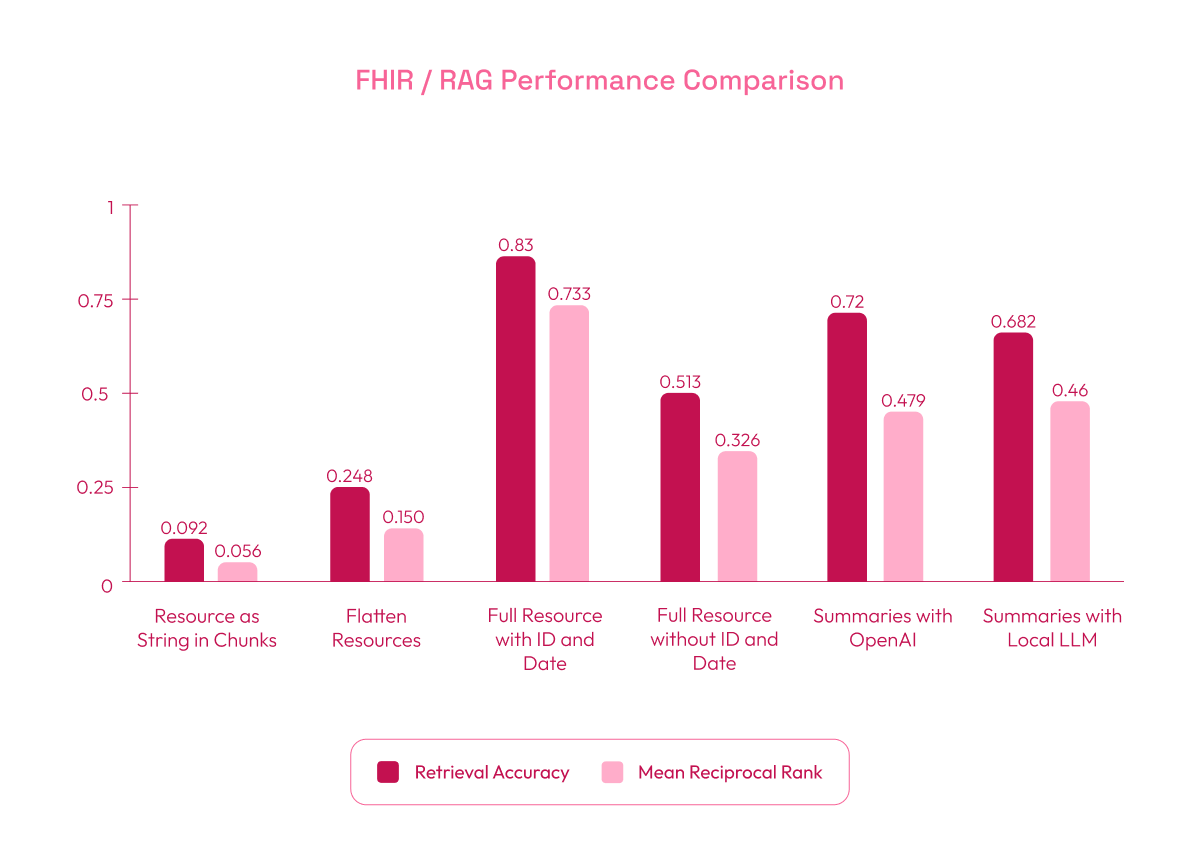
- Resource as String in Chunks Strategy (baseline): Performed poorly with a Retrieval Accuracy of 0.092 and MRR of 0.056. In this strategy, the evaluation dataset was generated by creating one question and answer per chunk, which led to excessive chunk segmentation and a loss of context. The overly simplified questions and fragmented data contributed to the low retrieval accuracy.
- Flatten Resources Strategy: Improved results, with a Retrieval Accuracy of 0.248 and MRR of 0.150. Flattening the data into simpler phrases enhanced retrieval by making the content easier to search.
- Full Resource as String with ID and Date: Achieved the best performance, with a Retrieval Accuracy of 0.83 and MRR of 0.733. Unlike the first approach, the evaluation questions and answers in this dataset were based on the complete resource rather than individual chunks. However, for storage in the vector database, the resources were still divided into chunks, but using the entire resource for question generation provided better context and improved retrieval accuracy. While adding IDs and dates helped reduce ambiguity and improved the retrieval of relevant data, we cannot expect all user queries to include such specific information. Instead, these values could be automatically inserted depending on the context in which the question is made, which is why we evaluated both scenarios.
- Full Resource String without ID and Date: Performance decreased to a Retrieval Accuracy of 0.513 and MRR of 0.326, as the search relied more on content without unique identifiers. The one-question-per-chunk approach still showed improvement over the initial setup.
- Summaries with OpenAI: Delivered strong results with a Retrieval Accuracy of 0.72 and MRR of 0.479. OpenAI’s concise summaries helped the system retrieve relevant information efficiently.
- Summaries with Local LLM (Phi 3.5 mini): Achieved a Retrieval Accuracy of 0.682 and MRR of 0.46, slightly lower than OpenAI but still competitive. This suggests that local models can be effective alternatives when the required hardware is available.
Future Optimizations
Future work could focus on:
- Replacing FHIR Codes: Substituting technical codes (e.g., LOINC) with full clinical descriptions will make the data more understandable for AI models, improving overall performance.
- Scraping External Data: Incorporating additional data from external links embedded in FHIR resources (such as diagnostic guidelines) can further enrich the AI’s responses.
- Integrating Knowledge Graphs: Knowledge graphs can model relationships between clinical data, helping the AI system understand complex queries and return more nuanced responses.
- Filtering Relevant Keys: Identifying and filtering out unnecessary keys from the FHIR data while keeping only those that are relevant can reduce noise and enhance the AI model's ability to focus on pertinent information.
Conclusion
By optimizing the retrieval and structuring of FHIR data, RAG systems in healthcare can provide clear explanations and address patient-specific questions, making complex medical information easier to understand.
These systems serve as a valuable complement to healthcare providers by helping patients interpret their lab results, diagnoses, and treatment options. This enhanced understanding not only improves communication between patients and specialists but also empowers patients to make informed decisions about their health.
Future advancements, such as integrating knowledge graphs and refining data descriptions, will further expand the capabilities of AI in healthcare, fostering a more informed and engaged patient experience.
We're passionate about pushing the boundaries of healthcare technology, constantly exploring ways to merge the latest research with practical solutions for our clients. If you're intrigued by this topic, or AI in healthcare, we'd love to connect. Whether you have questions, ideas to share, or are considering implementing similar solutions, don't hesitate to reach out!


